perEditor
A
tool to create personalized genome sequences
Home
Download
Installation
Usage
VCF
format
Tutorial
Overview
Usually
genomic reads, such as DNA-seq, RNA-seq, or ChIP-seq, are aligned
against a unique reference genome. However, mutation and recombination
processes produce variations from the reference sequence that can
mislead the alignment process. To overcome this problem, is necessary a
reference genome that has been modified to contain the variations of
the individual from which the samples were taken. Here, we created
perEditor, a reference genome editor that creates a copy of the
reference sequences that has been modified to reflex
information about SNPs and indels. Moreover, a second tool,
perEditor_ra, was created to be used on the previously
personalized reference sequences to take into account information
about chromosome rearrangements. Both perEditor and
perEditor_ra are command line tools that runs under Linux
systems.
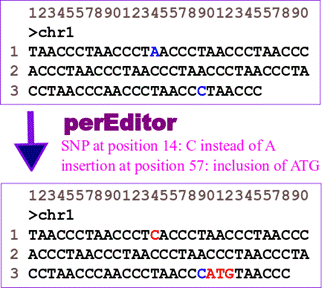
perEditor takes as input a fasta file for each one of the chromosomes of the reference genome, and a VCF file that contains information about SNPs and indels of such chromosome. Then, the tool creates a new fasta sequence that is a copy the original sequence, but that has been customized to contain the information contained on the VCF file. On the other hand, perEditor_ra can be used to further personalized the reference sequences. This time, chromosomal rearrangement information (provided in a VCF file) are used by perEditor_ra to manage the genetic variations described in the following figure.

Updates
|
6/14/2013
|
Versions of perEditor and perEditor_ra compiled under a 64-bites architecture are now available |
|
7 / 25 / 2011 |
perEditor_ra can handle chromosome inversions.
|
|
7 / 20 / 2011 |
A new tool, perEditor_ra, has been posted. perEditor_ra, allows the user to take into account chromosome rearrangement information in order to create personalized reference sequences |
Contact
Marcelo Rivas-Astroza ( rivasas2 AT Illinois DOT edu )